The Lab for AI in Medicine at TU Munich develops algorithms and models to improve medicine for patients and healthcare professionals.
Our aim is to develop artificial intelligence (AI) and machine learning (ML) techniques for the analysis and interpretation of biomedical data. The group focuses on pursuing blue-sky research, including:
- AI for the early detection, prediction and diagnosis of diseases
- AI for personalized interventions and therapies
- AI for the identification of new biomarkers and targets for therapy
- Safe, robust and interpretable AI approaches
- Privacy-preserving AI approaches
We have particularly strong interest in the application of imaging and computing technology to improve the understanding brain development (in-utero and ex-utero), to improve the diagnosis and stratification of patients with dementia, stroke and traumatic brain injury as well as for the comprehensive diagnosis and management of patients with cardiovascular disease and cancer.
The following research groups are based at the chair:
- AI for biomedical image analysis and interpretation
- Inverse problems in biomedical imaging
- Privacy-preserving and trustworthy AI in medicine
- AI for vision
- Artificial Intelligence for opportunistic cardiac MRI
Introduction
Medical imaging allows doctors to examine the interior structure or function of the human body often without the need for invasive surgical procedures. It comprises a range of different techniques, such as computed tomography (CT), magnetic resonance imaging (MR) and ultrasound (US). Clinicians rely on the information provided by medical imaging to monitor patients, diagnose illnesses and decide on treatment.
Our mission is to support doctors in the clinical process and improve patientcare by developing advanced algorithms that uses artificial intelligence (AI) techniques. To this end, we create and improve machine learning (ML) algorithms for various parts of the medical imaging pipeline. At the image level, we develop methods to tackle tasks such as segmentation of relevant anatomical structures, registration of images across time or modalities and enhancement of image quality. At the decision level, we innovate solutions to extract clinically useful information from medical images, diagnose diseases and predict future outcomes.
Challenges and Research Focuses
Developing these algorithms in the medical domain presents many challenges which we are striving to overcome. Medical data is often sparse and annotations for algorithm training are costly to acquire, with problems such as domain shift plaguing the few available data which could be detrimental to ML algorithms. For this, we are developing data efficient and domain robust solutions, as well as exploring opportunities provided by the increasing availability of large public datasets / biobanks.
In addition, medical images are usually accompanied by additional information from different sources such as doctor’s notes, laboratory test results or genomics data, all of which should be considered when interpreting the images. Part of our research centers on developing multi-modal AI solutions that integrate these diverse data sources, enabling more comprehensive image analysis.
Finally, to successfully deploy these algorithms in a hospital setting to improve patient care, it is essential to consider how they will be used by clinicians and how well they integrate into existing clinical workflows. This involves close collaboration with medical professionals in the hospital to align our research projects with clinical value, incorporating medical expertise into the development process, as well as improving interpretability of our ML algorithms to foster trust and facilitate adoption.
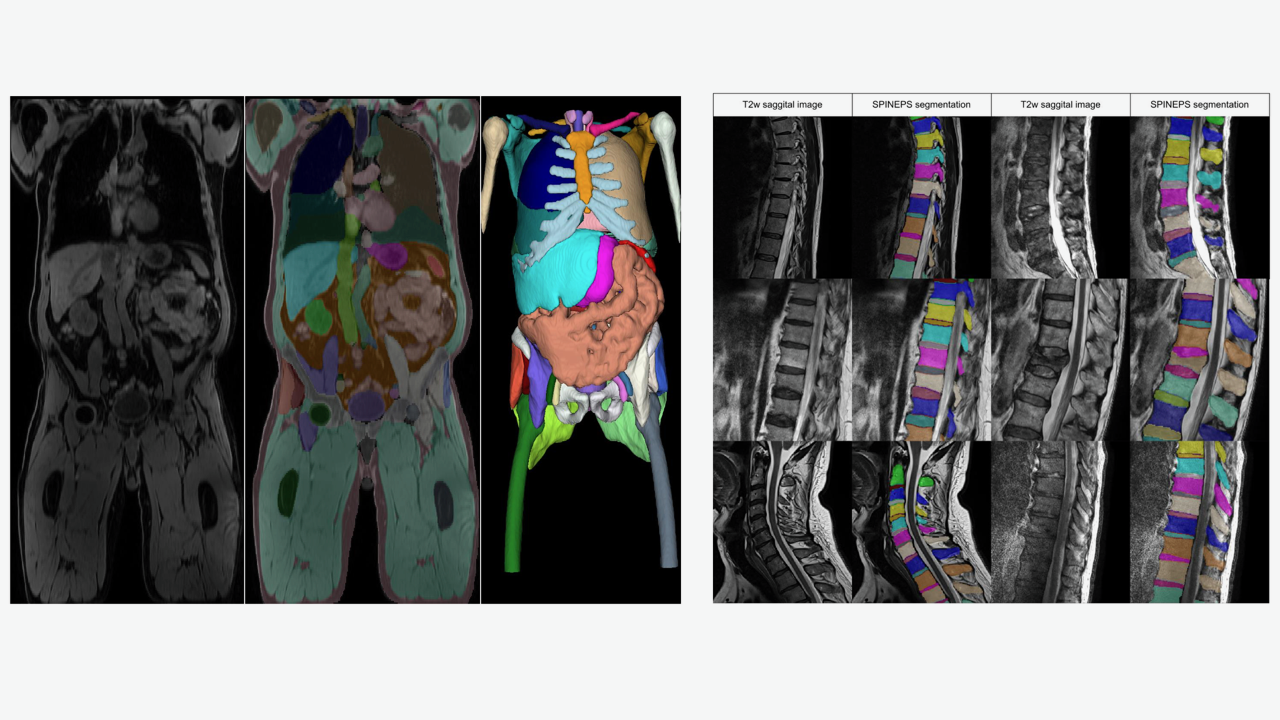
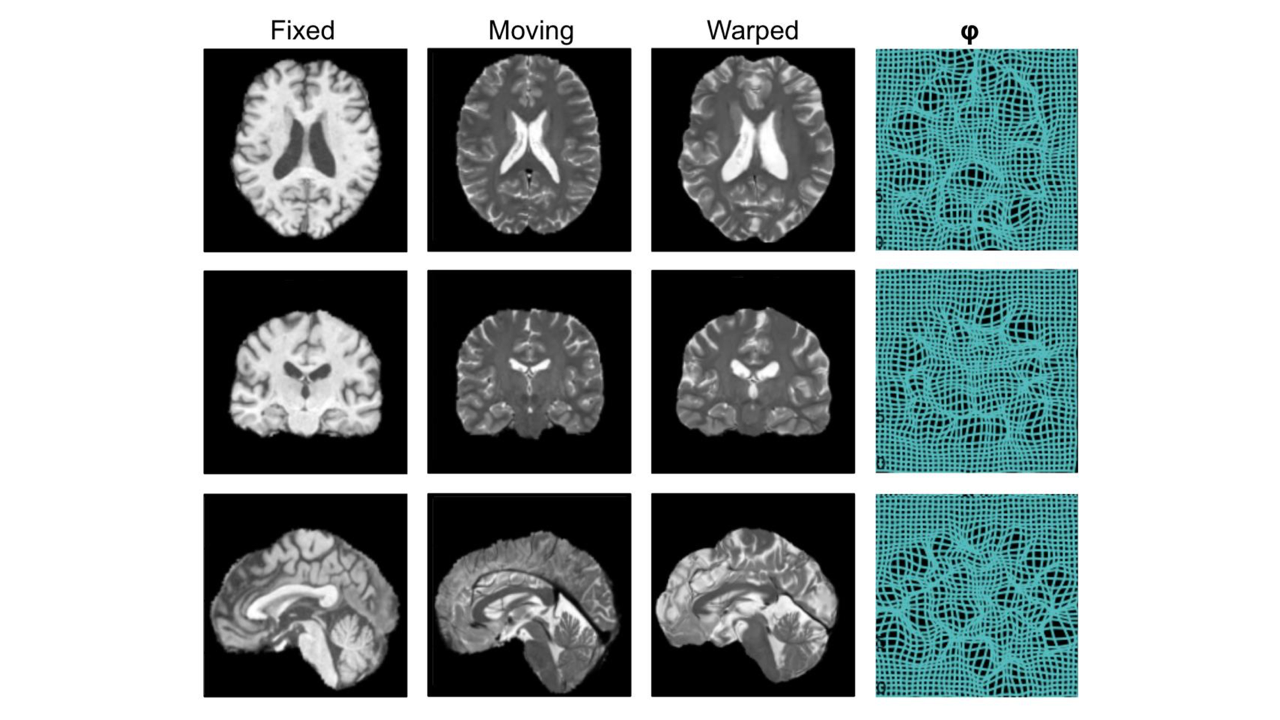
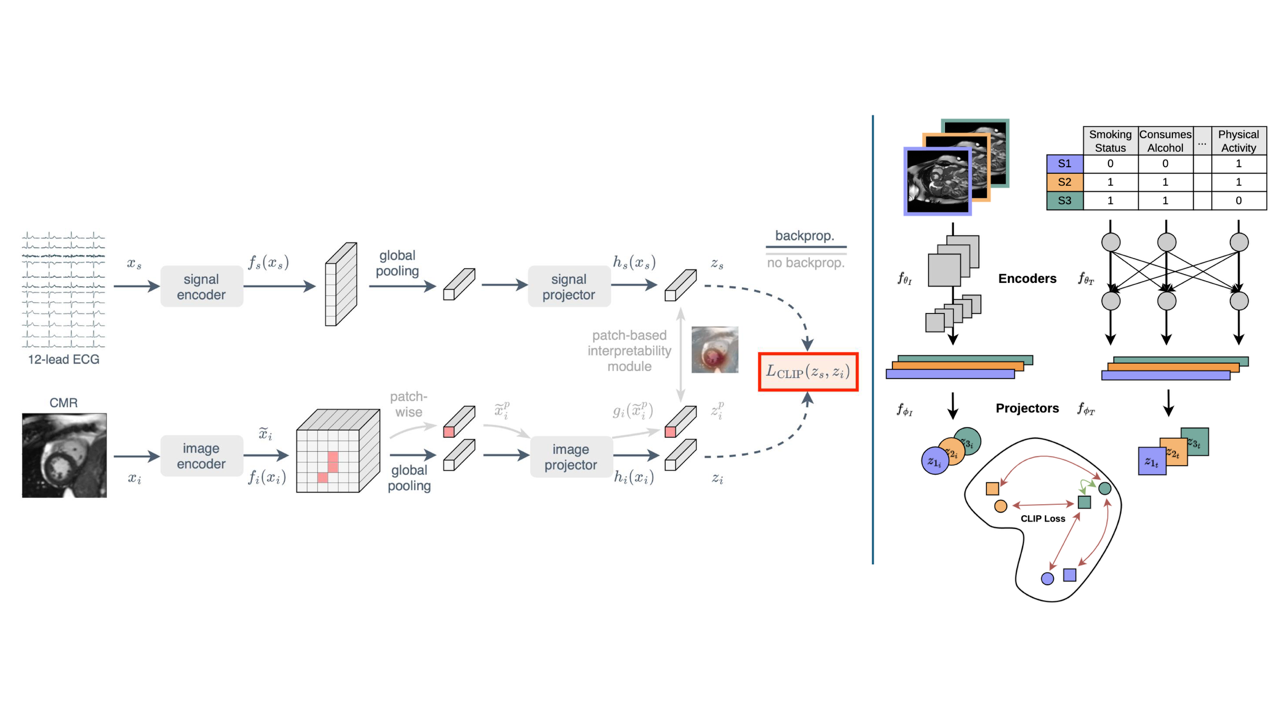
Key publications
- Hager, P., Jungmann, F., Holland, R., Bhagat, K., Hubrecht, I., Knauer, M.M., Vielhauer, J., Makowski, M., Braren, R., Kaissis, G., & Rueckert, D. (2024). Evaluation and mitigation of the limitations of large language models in clinical decision-making. Nature Medicine, 30, 2613 – 2622.
- Dima, A.F., Zimmer, V.A., Menten, M.J., Li, H.B., Graf, M., Lemke, T., Raffler, P., Graf, R., Kirschke, J.S., Braren, R.F., & Rueckert, D. (2023). 3D Arterial Segmentation via Single 2D Projections and Depth Supervision in Contrast-Enhanced CT Images. International Conference on Medical Image Computing and Computer-Assisted Intervention.
- Turgut, Ö., Müller, P., Hager, P., Shit, S., Starck, S., Menten, M.J., Martens, E., & Rueckert, D. (2023). Unlocking the Diagnostic Potential of ECG through Knowledge Transfer from Cardiac MRI.
- Müller, P., Kaissis, G., & Rueckert, D. (2024). ChEX: Interactive Localization and Region Description in Chest X-rays. European Conference on Computer Vision.
- Mueller, T.T., Starck, S., Bintsi, K., Ziller, A., Braren, R., Kaissis, G., & Rueckert, D. (2024). Are Population Graphs Really as Powerful as Believed? Trans. Mach. Learn. Res., 2024.
- Sideri-Lampretsa, V., McGinnis, J., Qiu, H., Paschali, M., Simson, W., & Rueckert, D. (2024). SINR: Spline-enhanced implicit neural representation for multi-modal registration. In Medical Imaging with Deep Learning.
- Berger, A.H., Stucki, N., Lux, L., Buergin, V., Shit, S., Banaszak, A., Rueckert, D., Bauer, U., & Paetzold, J.C. (2024). Topologically faithful multi-class segmentation in medical images. International Conference on Medical Image Computing and Computer-Assisted Intervention.
- Dannecker, M., Kyriakopoulou, V., Cordero-Grande, L., Price, A., Hajnal, J.V., & Rueckert, D. (2024). CINA: Conditional Implicit Neural Atlas for Spatio-Temporal Representation of Fetal Brains. International Conference on Medical Image Computing and Computer-Assisted Intervention.
- Starck, S., Sideri-Lampretsa, V., Ritter, J. J., Zimmer, V. A., Braren, R., Mueller, T. T., & Rueckert, D. (2024). Using UK Biobank data to establish population-specific atlases from whole body MRI. Communications Medicine, 4(1), 237.
- Zhang, Y., Chen, C., Shit, S., Starck, S., Rueckert, D., & Pan, J. (2024, October). Whole heart 3d+ t representation learning through sparse 2d cardiac mr images. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 359-369). Cham: Springer Nature Switzerland.
Introduction
Our group is working on inverse problems in biomedical imaging and their solution using artificial intelligence and machine learning.
Deep learning for inverse problems
The development of algorithms to solve inverse problems arising in sensor and imaging systems has a long tradition. Examples include compressed sensing approaches, e.g. for medical and computational imaging. Until recently, most algorithms for inverse problems were based on statistical or physical signal models, such as wavelets or sparse representations. Our research focuses on novel approaches based on deep learning to accelerate solving such problems.
AI-based image reconstruction
The focus of research here is how these deep learning-based approaches can be optimized for clinical applications and how they can be combined with image analysis methods. Deep learning-based approaches for reconstructing magnetic resonance imaging (MRI) or computed tomography (CT) provide efficient AI models, allowing the reconstruction of high-quality MRI images, and high-quality CT images from low-dose X-ray images.
For example, recent works on generative models have shown great promise for accelerating reconstruction tasks to shorten the scan time in MRI, as well as generating images with much higher resolution than the acquired resolution (e.g. super-resolution). Here, we tackle these problems by utilizing AI (i.e., diffusion models) guided by readily available MR images from other organs/tissues, MRI scanning parameters, or other MRI physics-guided information.
Key publications
- Schlemper, J., Caballero, J., Hajnal, J.V., Price, A.N. and Rueckert, D., 2017. A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE transactions on Medical Imaging
- Qin, C., Schlemper, J., Caballero, J., Price, A.N., Hajnal, J.V. and Rueckert, D., 2018. Convolutional recurrent neural networks for dynamic MR image reconstruction. IEEE transactions on medical imaging
- Hammernik, K., Schlemper, J., Qin, C., Duan, J., Summers, R.M. and Rueckert, D., 2021. Systematic evaluation of iterative deep neural networks for fast parallel MRI reconstruction with sensitivity‐weighted coil combination. Magnetic Resonance in Medicine
- Hammernik, K., Küstner, T., Yaman, B., Huang, Z., Rueckert, D., Knoll, F. and Akçakaya, M., 2023. Physics-Driven Deep Learning for Computational Magnetic Resonance Imaging: Combining physics and machine learning for improved medical imaging. IEEE signal processing magazine
- Huang, W., Li, H.B., Pan, J., Cruz, G., Rueckert, D. and Hammernik, K., 2023, June. Neural implicit k-space for binning-free non-cartesian cardiac MR imaging. In International Conference on Information Processing in Medical Imaging
- Pan, J., Hamdi, M., Huang, W., Hammernik, K., Kuestner, T. and Rueckert, D., 2024. Unrolled and rapid motion-compensated reconstruction for cardiac CINE MRI. Medical Image Analysis
- Pan, J., Huang, W., Rückert, D., Küstner, T. and Hammernik, K., 2024. Reconstruction-driven motion estimation for motion-compensated MR CINE imaging. IEEE Transactions on Medical Imaging
- Ezhov, I., Scibilia, K., Giannoni, L., Kofler, F., Iliash, I., Hsieh, F., Shit, S., Caredda, C., Lange, F., Montcel, B., Tachtsidis, I., Rueckert, D., 2024. Learnable real-time inference of molecular composition from diffuse spectroscopy of brain tissue. Journal of Biomedical Optics
- Chung, H., Lee, D., Wu, Z., Kim, B. H., Bouman, K. L., & Ye, J. C. (2025). ContextMRI: Enhancing Compressed Sensing MRI through Metadata Conditioning. arXiv preprint arXiv:2501.04284.
- Jiang, L., Mao, Y., Wang, X., Chen, X., & Li, C. (2023, October). Cola-diff: Conditional latent diffusion model for multi-modal mri synthesis. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 398-408). Cham: Springer Nature Switzerland.
- Rombach, R., Blattmann, A., Lorenz, D., Esser, P., & Ommer, B. (2022). High-resolution image synthesis with latent diffusion models. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 10684-10695).
Introduction
Our group is developing the next generation of privacy-preserving, secure, and trustworthy AI algorithms for medical applications.
Artificial Intelligence: Privacy, Security, and Safety
AI in medicine requires large, diverse, and representative datasets to train fair, generalizable, and reliable models. However, such datasets often contain sensitive personal information. Privacy-preserving machine learning bridges the gap between data utilization and data protection by enabling the training of AI models on private data while providing formal privacy guarantees.
Our group focuses on:
- Differential privacy (DP) theory and applications to machine learning and deep learning, targeting both unstructured datasets (e.g., images) and structured data (e.g., tabular and graph databases). This includes developing new theoretical frameworks for DP as well as practical implementations that balance privacy guarantees with model utility.
- Generative models and their applications, such as large language models (LLMs) and agentic systems that utilize them, integrating differential privacy to ensure secure and privacy-preserving outcomes.
- Data attribution techniques, which enable transparent and accountable data usage in training and inference, crucial for both privacy and intellectual property concerns.
- Developing techniques to mitigate trade-offs between privacy, model utility, and computational efficiency.
- AI security, including the study of vulnerabilities in collaborative machine learning protocols (e.g., federated learning) and designing robust defense mechanisms against adversarial attacks.
Artificial Intelligence and Trustworthiness
Building trust in AI necessitates a comprehensive approach encompassing privacy, reliability, and security. Our group works on trustworthy machine learning foundations, which include:
- Quantifying uncertainty in model outputs to make predictions interpretable and reliable.
- Incorporating domain expertise to ensure models align with expert expectations and context-specific requirements.
- Developing probabilistic machine learning models to counteract poorly calibrated predictions from conventional models.
- Employing computational Bayesian techniques to train statistical and deep learning algorithms.
- Exploring the intersection of probabilistic and privacy-preserving machine learning, ensuring robust models that are secure, transparent, and fair.
AI Safety and Responsible Generative Systems
As AI systems increasingly integrate generative models like LLMs, their safe deployment becomes paramount. Our group actively works on:
- Establishing formal guarantees of safety and reliability for agentic LLM applications.
- Exploring robustness in generative systems to reduce risks associated with misuse or erroneous outputs.
- Ensuring the alignment of generative models with human values and ethical guidelines, particularly in high-stakes domains like healthcare.
Through this multifaceted focus on privacy, security, and trust, we aim to set a standard for AI that is not only powerful but also safe, ethical, and respectful of individual rights.
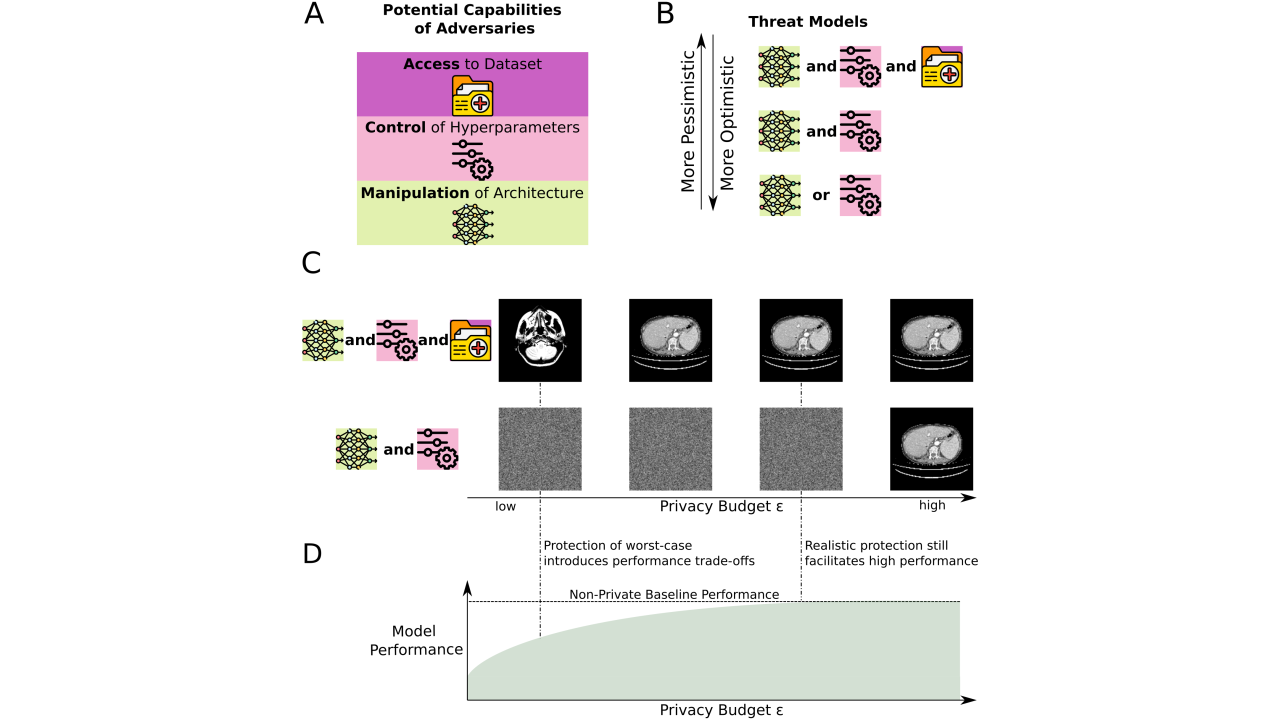

Key publications
- Ziller, A., Mueller, T., Stieger, S., Feiner, L., Brandt, J., Braren, R., Rueckert, D., Kaissis, G., 2024. Reconciling Privacy and Accuracy in AI for Medical Imaging. Nature Machine Intelligence
- Hager, P., Jungmann, F., Holland, R., Bhagat, K., Hubrecht, I., Knauer, M., Vielhauer, J., Makowski, M., Braren, R.*, Kaissis, G.*, Rueckert, D.* (*equal contribution), 2024. Evaluating and Mitigating Limitations of Large Language Models in Clinical Decision Making. Nature Medicine
- Kaissis, G., Kolek, S., Balle, B., Hayes, J. and Rueckert, D., 2024. Beyond the calibration point: Mechanism comparison in Differential Privacy. International Conference on Machine Learning
- Tayebi Arasteh, S., Ziller, A., Kuhl, C., Makowski, M., Nebelung, S., Braren, R., Rueckert, D., Truhn, D. and Kaissis, G., 2024. Preserving fairness and diagnostic accuracy in private large-scale AI models for medical imaging. Communications Medicine
- Kaissis, G., Ziller, A., Kolek, S., Riess, A. and Rueckert, D., 2023, Optimal privacy guarantees for a relaxed threat model: Addressing sub-optimal adversaries in differentially private machine learning., Advances in Neural Information Processing Systems (NEURIPS)
- Raab, R., Küderle, A., Zakreuskaya, A., Stern, A.D., Klucken, J., Kaissis, G., Rueckert, D., Boll, S., Eils, R., Wagener, H. and Eskofier, B.M., 2023. Federated electronic health records for the European Health Data Space. The Lancet Digital Health
- Meiser, P., Knolle, M., [...], Kaissis, G.* and Böttcher, J.* (*equal contribution), 2023. A distinct stimulatory cDC1 subpopulation amplifies CD8+ T cell responses in tumors for protective anti-cancer immunity, Cancer Cell
- Mueller, T.T., Paetzold, J.C., Prabhakar, C., Usynin, D., Rueckert, D. and Kaissis, G., 2022. Differentially Private Graph Neural Networks for Whole-Graph Classification. IEEE Transactions on Pattern Analysis and Machine Intelligence (TPAMI)
- Usynin, D., Ziller, A., Makowski, M., Braren, R., Rueckert, D., Glocker, B., Kaissis, G. and Passerat-Palmbach, J., 2021. Adversarial interference and its mitigations in privacy-preserving collaborative machine learning. Nature Machine Intelligence
- Kaissis, G., Ziller, A., Passerat-Palmbach, J., Ryffel, T., Usynin, D., Trask, A., Lima Jr, I., Mancuso, J., Jungmann, F., Steinborn, M.M. and Saleh, A., 2021. End-to-end privacy preserving deep learning on multi-institutional medical imaging. Nature Machine Intelligence
- Dou, Q., So, T.Y., Jiang, M., Liu, Q., Vardhanabhuti, V., Kaissis, G., Li, Z., Si, W., Lee, H.H., Yu, K. and Feng, Z., 2021. Federated deep learning for detecting COVID-19 lung abnormalities in CT: a privacy-preserving multinational validation study. NPJ Digital Medicine
- Kaissis, G., Makowski, M.R., Rückert, D. and Braren, R.F., 2020. Secure, privacy-preserving and federated machine learning in medical imaging. Nature Machine Intelligence
Introduction
The AI for Vision group focuses on blue-sky research in medical image analysis with a particular focus on the application of machine learning and computer vision algorithms in the field of ophthalmology. Specifically, we are working on:
Self-supervised learning
Labeling medical data is very expensive, as it is time-consuming and requires expert knowledge. Moreover, medical data often includes highly sensitive information, making it challenging to share without compromising the privacy of the subjects involved. To overcome the limited availability of large annotated medical datasets, we are researching self-supervised learning. This approach leverages unlabeled medical data to enable neural networks to extract meaningful features that can be effectively adapted to a wide range of downstream tasks.
Multimodal deep learning
Clinicians rarely rely on a single source of information when diagnosing patients and deciding on a course of action. In order to place their observations into context, they consider an array of multimodal data, such as demographic and genomic information, patient interviews, laboratory test results and biomedical images. As the number of diagnostic modalities grows and the complexity of the obtained data increases, comprehensively evaluating all pieces of information is becoming more and more challenging for healthcare professionals. To address this, our research focuses on developing deep learning algorithms capable of integrating diverse multimodal data to support autonomous and effective clinical decision making.
Deep learning for ophthalmology
Good vision is essential for navigating our environment, communicating, and performing everyday activities, such as reading, exercising, and working. Conversely, low vision or blindness severely impacts both the physical and emotional well-being of patients. As of 2020, more than 200 million people worldwide suffered from moderate to severe vision impairment. Research and treatment of eye diseases is increasingly being driven by sophisticated imaging modalities and advanced image processing methods. Driven by the comparative ease to image the eye and obtain large imaging datasets, ophthalmology has been an early adopter of deep learning in healthcare. Our group’s work in machine learning for ophthalmology simultaneously evaluates new algorithmic innovations while aiming to improve medical care for patients affected by ocular diseases.
Key publications
- R. Holland, O. Leingang, H. Bogunović, S. Riedl, L. Fritsche, T. Prevost, H. P. N. Scholl, U. Schmidt-Erfurth, S. Sivaprasad, A. J. Lotery, D. Rueckert, and M. J. Menten. “Metadata-enhanced contrastive learning from retinal optical coherence tomography images.” Medical Image Analysis, 97:103296, 2024
- L. Kreitner, J. C. Paetzold, N. Rauch, C. Chen, A. M. Hagag, A. E. Fayed, S. Sivaprasad, S. Rausch, J. Weichsel, B. H. Menze, M. Harders, B. Knier, D. Rueckert, and M. J. Menten. “Synthetic opticalcoherence tomography angiographs for detailed retinal vessel segmentation without humanannotations.” IEEE Transactions on Medical Imaging, 43(6):2061–2073, 2024
- M. J. Menten, J. C. Paetzold, V. A. Zimmer, S. Shit, I. Ezhov, R. Holland, M. Probst, J. A. Schnabel, and D. Rueckert. “A skeletonization algorithm for gradient-based optimization.” Proceedings of the IEEE/CVF International Conference on Computer Vision, 21394–21403, 2023
- R. Holland, O. Leingang, C. Holmes, P. Anders, R. Kaye, S. Riedl, J. C. Paetzold, I. Ezhov, H.Bogunović, U. Schmidt-Erfurth, H. P. N. Scholl, S. Sivaprasad, A. J. Lotery, D. Rueckert, and M. J. Menten. “Clustering disease trajectories in contrastive feature space for biomarker proposal in age-related macular degeneration.” International Conference on Medical Image Computing and Computer-Assisted Intervention, 724–734. 2023
- M. J. Menten, R. Holland, O. Leingang, H. Bogunović, A. M. Hagag, R. Kaye, S. Riedl, G. L. Traber, O. N. Hassan, N. Pawlowski, B. Glocker, L. G. Fritsche, H. P. N. Scholl, S. Sivaprasad, U. Schmidt- Erfurth, D. Rueckert, and A. J. Lotery. “Exploring healthy retinal aging with deep learning.” Ophthalmology Science, 3(3):100294, 2023
- P. Hager, M. J. Menten, and D. Rueckert. “Best of both worlds: Multimodal contrastive learning with tabular and imaging data.” Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 23924–23935, 2023
- M. J. Menten, J. C. Paetzold, A. Dima, B. H. Menze, B. Knier, and D. Rueckert. “Physiology-basedsimulation of the retinal vasculature enables annotation-free segmentation of OCT angiographs.”International Conference on Medical Image Computing and Computer-Assisted Intervention, 330–340, 2022